Kinesin nomenclature
Andreas Prokop raised the issue of kinesin nomenclature on twitter over the weekend “I am tired of having to look up lists!”. I couldn’t agree more, it’s insanely confusing - especially when you’re trying to synthesise literature across several model organisms (shout out to Ustilago maydis and Dictyostelium discoideum here). Why is it confusing? The standardised kinesin nomenclature was defined in 2004, where the superfamily was divided into sub families. They even defined how motors should be referred to in the text.
- Human Eg5, a member of the Kinesin-5 family (previously referred to as BimC by Dagenbach and Endow, 2004), is involved in establishing the bipolar spindle.
- Human Eg5, a member of the Kinesin-5 family, is involved in establishing the bipolar spindle.
The issue is that this is a mouthful (plus words & characters), so what tends to happen is people stick to the gene/protein name and you have to google to figure out sub-family before you can figure out how it might relate to your own system. So we’re not really talking about nomenclature, just an efficient way of getting from (slightly random) protein/gene name to sub-family information. Preferably without increasing our words counts.
Andreas proposed superscript to solve the problem, but this definitely has problems too:
We need better kinesin nomenclature. What about Kin1[KIF5a], Kin6[KIF20a], Kin6[pav] (with square brackets being superscript) - I am tired of having to look up lists! - image: Suppl. Mat. of https://t.co/77GWj7rZgzhttps://t.co/idk0D8bgQz pic.twitter.com/MNS2ZsecTD
— Andreas Prokop (@Poppi62) December 10, 2017
I don't like superscripts because they're difficult to write with something other than Word, hard to read, lost when copy/pasting. What's wrong with the slash KIF5/kinesin-1?
— Christophe🔬Leterrier (@christlet) December 10, 2017
So what’s the solution? I mentioned something I’d adopted when trying to define subpopulations of the dynein motor.
I agree it doesn’t work right now. For my part I use kinesin-1 referring to KIF5 + KLC. I’ve recently stolen from R syntax to describe dynein defined by subunit inclusion as dynein::DIC1 and dynein::DIC2. Could work?
— Ali Twelvetrees (@dozenoaks) December 10, 2017
I think it’s worth thinking about syntax like this, because it’s a structured way of tackling the problem. When I started using :: to define dynein::DIC1 and dynein::DIC2 in my own notes it happened naturally without me really thinking about it, then stuck. I took the :: operator from R where it disambiguates functions with the same name from different packages. I was thinking about dynein function being defined by certain subunits so dynein::DIC1 felt natural, meaning (in my head) the dynein function defined by incorporation of the DIC1 subunit. Some dynein subunits act outside of dynein in other complexes (packages), so you can imagine dynein::LC8 and some_other_complex::LC8.
I don’t think the kinesin problem is similar to this though. Defining kinesin sub-types is more like calling from a list. There is the kinesin-1 sub-family that has many members with different names across species, and you’re referring to one member. Calling from a list in R is straightforward:
#' First I bulid a list
kinesin_1 <- list(KIF5A = "kinesin-1 isoform KIF5A",
KIF5B = "kinesin-1 isoform KIF5B",
KIF5C = "kinesin-1 isoform KIF5C")
#' Then I can call from the list
#' First the whole list
kinesin_1# $KIF5A
# [1] "kinesin-1 isoform KIF5A"
#
# $KIF5B
# [1] "kinesin-1 isoform KIF5B"
#
# $KIF5C
# [1] "kinesin-1 isoform KIF5C"#' Now just the bit I'm interested in
kinesin_1[1]# $KIF5A
# [1] "kinesin-1 isoform KIF5A"I don’t think using the $ in the middle of a manuscript will catch on. So, maybe as suggested by Andreas, the square brackets are a good compromise? Kinesin_1[KIF5A] being the same as kin1[KIF5A]. The test: the kinesin-1 motor in Ustilago maydis is called kin2, so will we know what kin1[kin2] means relative to kin2[kin2]?
Actually, the Kin1/KIF5a solution feels optional. I would then prefer Kin[KIF5] or Kin1:KIF5 (two colons would indicate a fusion protein, wouldn't they? @dozenoaks @bengoult @antoine_triller @clathrin https://t.co/tzpfXUmNDb
— Andreas Prokop (@Poppi62) December 11, 2017
(to be continued…?)
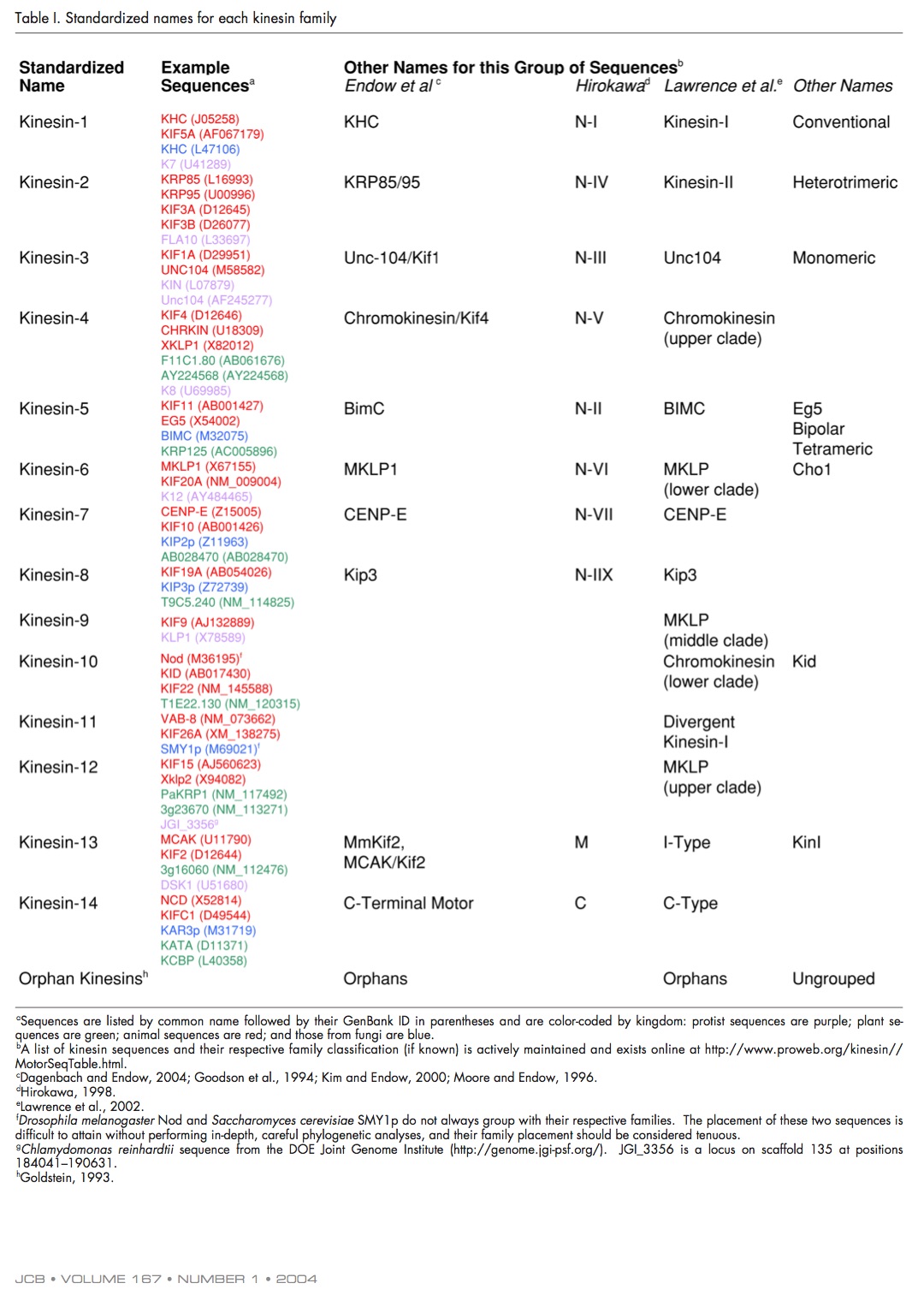
Fig 1. Kinesin nomenclature from Lawrence et al., 2004
